Targeted Protein Degradation
What is targeted protein degradation?
Targeted protein degradation is an emerging therapeutic strategy for the modulation of pathogenic proteins by hijacking the cell’s own degradation machinery. This approach has expanded the druggable proteome space to challenging targets that were previously believed to be undruggable.
How does it work?
Eukaryotic cells have two primary pathways for disposing of unwanted proteins: the ubiquitin-proteasome pathway and the autophagy-lysosome pathway. The former generally removes short-lived and soluble misfolded proteins, while the latter removes long-lived proteins, insoluble protein aggregates, and even organelles via endocytosis, phagocytosis or autophagy. Targeted protein degraders act by hijacking one of these pathways and most degraders discovered to date function through the Ubiquitin-Proteasome System (UPS). These molecules act by forming a stable ternary complex with a ubiquitin ligase (E3) and an intracellular protein of interest (POI). As a result, the proteins are brought into proximity, and ubiquitin’s are added to the POI sequentially, forming a chain. The polyubiquitin chains can have different architecture and resulting effects on the fate and function of the targeted protein but typically lysine 48-linked polyubiquitin chains are desired for targeted protein degradation by the proteasome. Targeted protein degraders that use the lysosome-dependent pathway are not as common, but there have been successful examples of molecules capable of hijacking the endocytosis or autophagy pathways to degrade unwanted proteins of interest. As with the UPS strategy, these molecules act by establishing a ternary complex with a POI (either membrane-bound or extracellular) and a protein involved in the lysosome pathway. This is a new area of research that will surely see further development as it will open the door to the degradation of membrane proteins, extracellular proteins and organelles.
Targeted protein degraders
Several mechanisms have been researched for TPD and can be categorized based on their pathway and functional mechanism.
Targeted protein degraders via UPS:
- PROteolysis TArgeting Chimeras (PROTAC®)
- Molecular glues
- Specific and non-genetic inhibitors of apoptosis protein-dependent protein erosive agents (SNIPER)
- Hydrophobic Tags (HyT)
- Selective androgen receptor degrader (SARD)
- Selective estrogen receptor degrader (SERD)
- Transcriptor factor PROteolysis TArgeting Chimeras (TF-PROTAC)
Targeted protein degraders via the endosome-lysosome system:
- Lysosome-targeting Chimeras (LYTAC)
- Bispecific aptamer Chimera (BIAC)
- Antibody-based PROTAC (AbTAC)
- GlueTAC
Targeted protein degraders via autophagy-lysosome system:
- Autophagy-targeting Chimeras (AUTAC)
- Autophagosome-tethering compound (ATTEC)
- AUTOphagy TArgeting Chimera (AUTOTAC)
- CMA-based degrader
The field of targeted protein degraders has grown significantly since its inception in 1999. The most advanced targeted protein degraders in the clinic are PROTACs with currently several small molecules in phase II and phase III clinical trials and many more in drug discovery pipelines. Molecular glues are also gathering a lot of attention but are still at an earlier stage, despite some molecules being already in the clinic (their functional mechanisms were unknown at the time of the approval). The other protein targeted degraders described above were only recently identified, but they offer a complement to PROTACs and molecular glues for targeting extracellular proteins, membrane proteins, and organelles.
PROTACs & Molecular Glues vs traditional protein inhibition
PROTACs and molecular glues operate through mechanisms that are fundamentally different from traditional protein inhibition. While they promote the degradation of a POI, traditional occupancy-driven pharmacology acts by blocking the function of a protein. As a result, PROTACs and molecular glues have several advantages over traditional approaches, such as:
- Expansion of the druggable proteome to proteins that lack traditional ligandable sites.
- Elimination of physiological function of a disease-causing POI.
- Reduction of the likelihood of drug resistance from over-expression and mutation in long-term selection pressure. This is particularly important in traditional kinase inhibitors.
- Expected lower toxic side effects as PROTACs and molecular glues act in sub-stoichiometric/catalytic manner.
PROTACs vs Molecular Glues
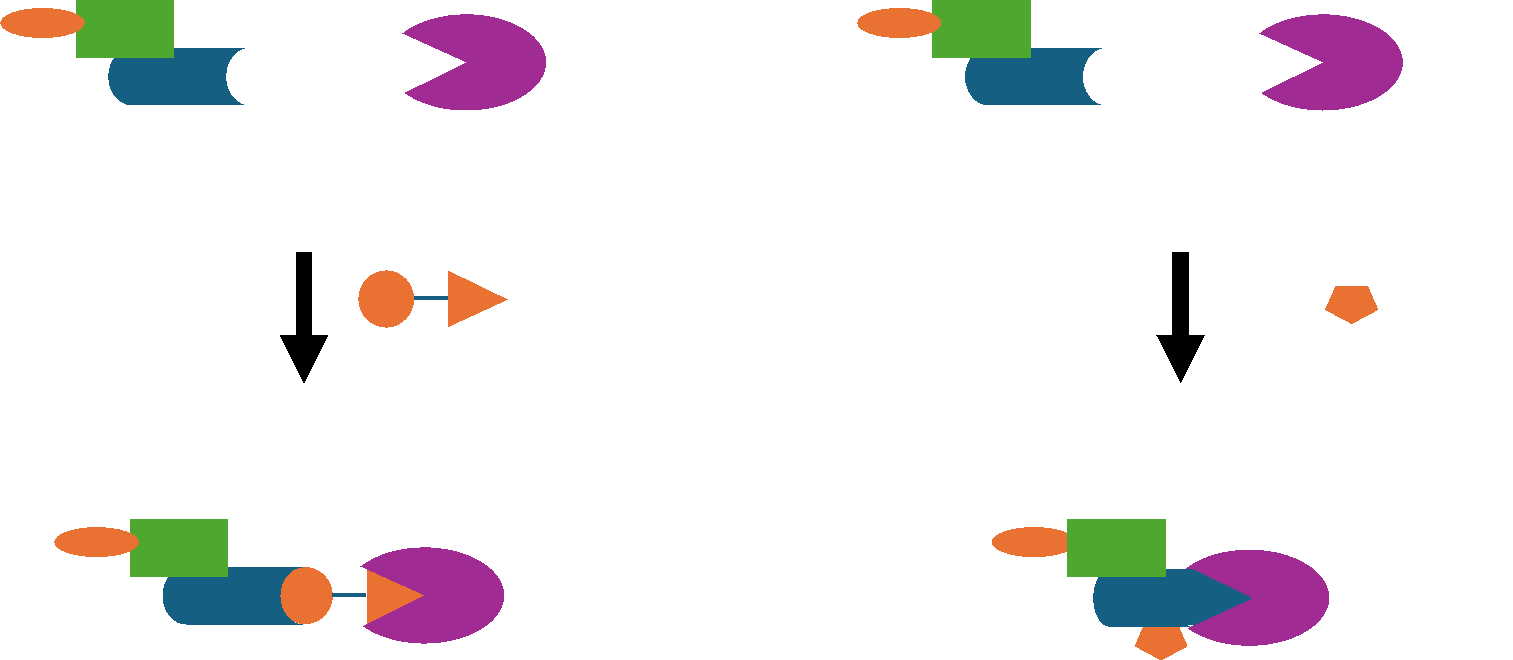
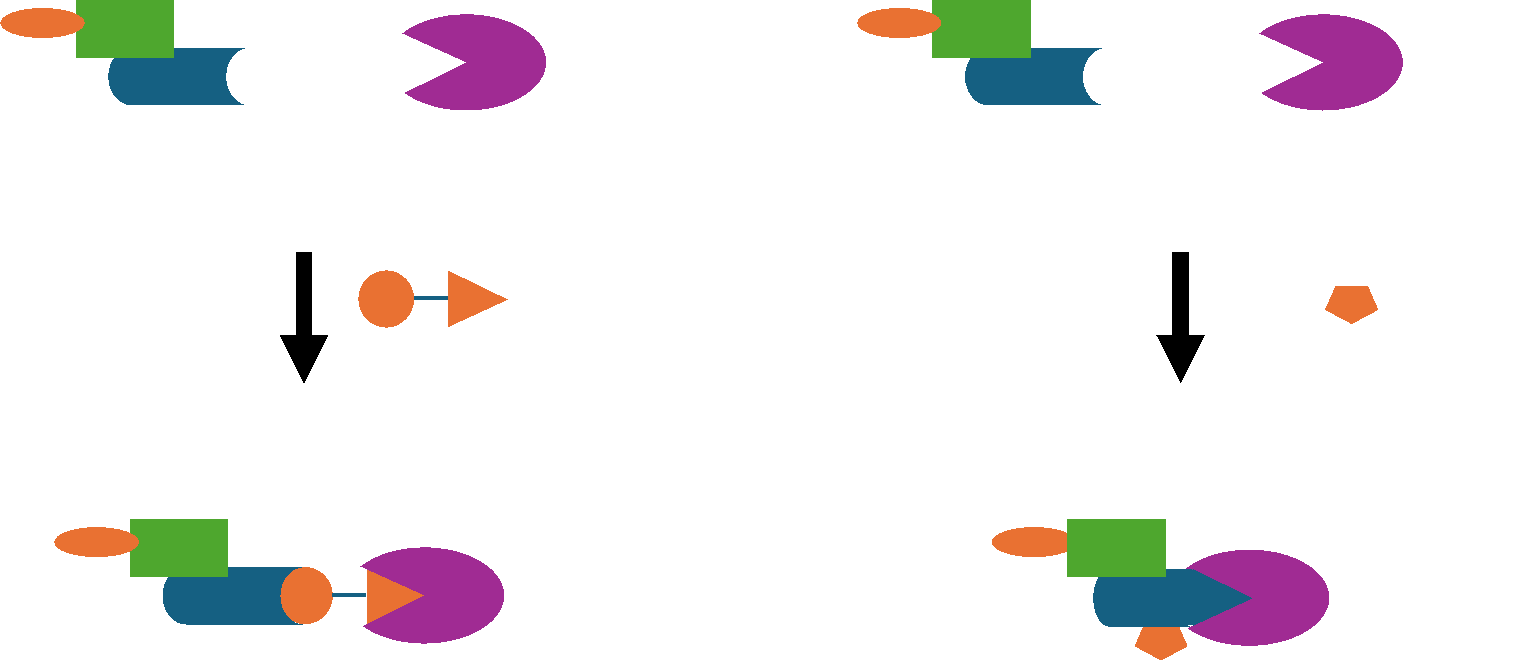
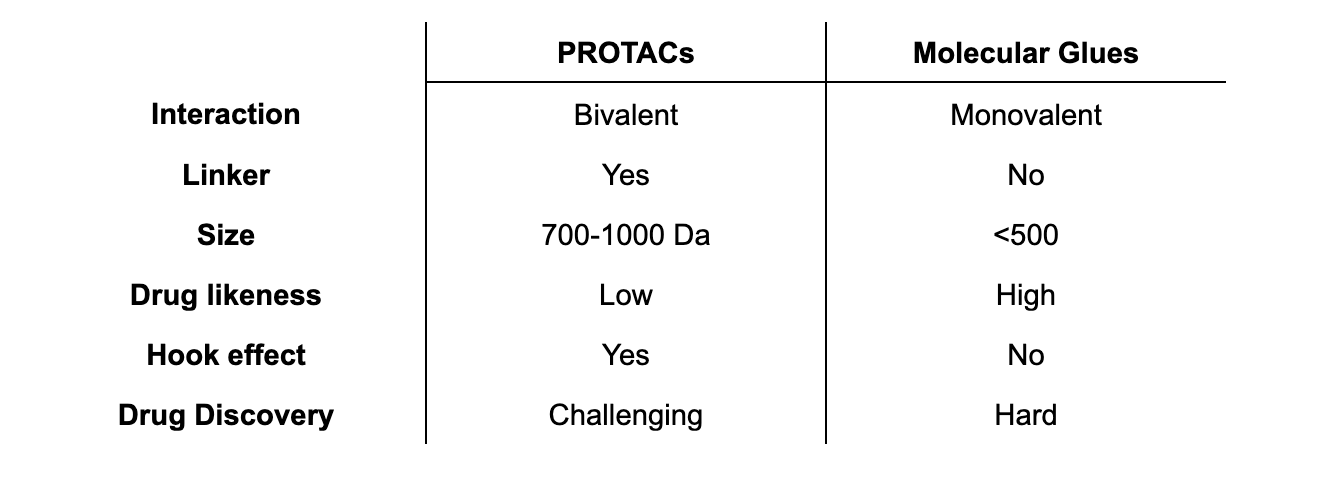
PROTACs and molecular glues act via the same mechanism, but they are notably different. PROTACs are bivalent molecules comprised of 3 different parts: i) a POI-targeting binding ligand, ii) an E3-recruiting warhead and iii) a flexible linker that connects the 2 ligands. PROTACs interact simultaneously with both proteins, thus promoting the ubiquitylation of the POI.
Conversely, molecular glues are monovalent compounds that interact primarily with either the ligase or the POI to increase the affinity between the two proteins, thereby also promoting the ubiquitylation and subsequent degradation of the POI.
PROTACs are large molecules that range between 700 to 1000 Da and therefore they do not comply to Lipinski’s rule-of-five. This means that oral bioavailability and cell permeability can be a challenge for these molecules. Thus, the chem-phys properties of PROTACs can constrain the route of administration and pose challenges during the hit-to-lead optimization. Moreover, an underlying characteristic of all PROTACs is the ability to form binary complex with either the POI or the E3 ligase when present in excess concentrations. This is the so-called hook effect and can reduce the target degradation but also lead to degradation of neo-substrates. Despite all challenges, PROTACs are versatile tools that allow for a modular design where one E3 ligase can be connected to many targets, making PROTAC development a much-used avenue in drug discovery.
Molecular glues are emerging as a new and promising therapeutic alternative to PROTACs. Not only do they have the same overall mechanism of action as PROTACs, but they have a better pharmacological profile. As simpler, monofunctional molecules without linkers, molecular glues are smaller on average and more easily satisfy Lipinski’s rule-of-five, an important factor in developing orally bioavailable drugs. Moreover, molecular glues can open the door of targeted protein degraders to CNS disorders, a field that is unlikely to become accessible to PROTACs as they are too big to cross the blood brain barrier. Secondly, as the binding is monovalent, the Hook effect is not observed at high concentrations of the ligand, contributing to an increase in efficacy and safety.
Hit identification in PROTACs and Molecular Glues
The drug discovery of PROTACs is complex and differs significantly from traditional approaches. It can be roughly divided in 4 stages:
- Hit identification – find a molecule that strongly binds to a POI, preferably in the nM range. In contrast with classical drug discovery, for a PROTAC to be effective, the molecule should just bind selectively to the POI, with no requirement on whether it exerts a modulating effect.
- E3 ligase evaluation – there are >600 E3 ubiquitin ligases encoded in the human genome. The selection of E3 ligase binding moieties is usually driven by physicochemical constraints and E3 ligase expression profiles of the targeted cells. Further, the E3 ligase binding moiety should not inhibit the function of the E3 ligase.
- Linker attachment and optimization – the hit should contain an attachment point to a linker. This stage is challenging as the location of the linker should not disrupt the binding. Molecules identified in DNA Encoded Library (DEL) screening have an advantage here, since discovered molecules are linked to DNA during the screening. Long and flexible linkers are initially incorporated to determine whether the POI can be degraded by the E3 ligase. Once the proof-of-concept is established, the linker is optimized for function and flexibility and physicochemical properties.
As molecular glues are monovalent and linker-less, the drug discovery approach is distinct from the discovery approach taken for PROTACs. The most notorious molecular glue degraders were in fact only retrospectively discovered to act as such. For example, thalidomide and analogues, now referred as immunomodulatory imide drugs (IMiDs), were approved by FDA and used for cancer treatment long before it was discovered that the E3 ligase cereblon was their target. Another example is indisulam, identified in a classical phenotypic screening and 20 years later discovered to be a molecular glue having RBM39 as its degradation target and DCAF15 as the E3 ligase effector. These two poster cases underscore the pharmacological potential of molecular glues but also emphasizes the need for new drug screening tools as the molecular glues were identified serendipitously.
Currently, more than 600 E3 ligases have been reported, but only five have been used for molecular glue-mediated degradation, namely CRBN, DDB1, β-TrCP, DCAF15, and SIAH1. Apart from the identification of new molecular glues tagging into these well-known E3 ligases, the plethora of E3 ligases available holds promise for massive progress as the full potential of the E3 ligase family unfolds.
Recent advances in hit identification techniques, including high-throughput screening (HTS), DNA-encoded libraries, structure-based drug design, data mining, and mass spectrometry, are expected to accelerate progress in this field significantly
See more about Vipergen’s solutions for
Discovery of molecular glues by clicking here
Discovery of binders, modulators and binders in living cells by clicking here
Discovery of binders, modulators and inhibitors in plastic by clicking here
References
- Zhao, L., Zhao, J., Zhong, K. et al. Targeted protein degradation: Mechanisms, strategies and application. Sig Transduct Target Ther 2022 7, 113.
- Zhong, G., Chang, X., Xie, W. et al. Targeted protein degradation: advances in drug discovery and clinical practice. Sig Transduct Target Ther 2024 9, 308.
- Paudel, R.R., Lu, D., Chowdhury, S.R., et al. Targeted Protein Degradation via Lysosomes. Biochemistry. 2023 February 07; 62(3): 564.
- Sincere, N.I., Anand, K., Ashique, S., et al. PROTACs: Emerging targeted protein degradation approaches for advanced druggable strategies.
Molecules 2023, 28, 4014. - Shen, F. and Dassama, L.M.K., Opportunities and challenges of protein-based
targeted protein degradation Chem. Sci., 2023, 14, 843 - Ogawa, Y.; Ueda, T.P.; Obara, et al., targeted protein degradation systems: Controlling protein stability using E3 ubiquitin ligases in eukaryotic species. Cells 2024, 13, 175
- Yoon, H., Rutter, J.C., Li, Y.-D. and Ebert, B.L. Induced protein degradation for therapeutics: Past, present, and future, J Clin Invest., 2024, 134, 1
PROTAC® is a registered trademark of Arvinas Operations, Inc.