DEL – Molecular Glue Direct – Tap into the potential of molecular glue
Molecular glues in targeted protein degradation
Unlocking the potential of targeted protein degradation (TPD) through the cellular ubiquitin-proteasome machinery represents a paradigm shift in drug discovery, broadening the scope of druggable targets. This process involves stabilizing a complex between the protein of interest (POI) and one of the 600+ E3 ubiquitin ligases encoded by the human genome, promoting POI ubiquitination and subsequent degradation.
The TPD field has been spearheaded by heterobifunctionals, such as PROTACS, whereas only a limited set of molecular glues have been discovered and advanced. Heterobifunctionals are easier to discover but more difficult to advance due to their size/physiochemical properties and the “hook effect”. In contrast molecular glues are more difficult to discover but easier to advance due to their better physicochemical properties and mode of action.
Our unique DEL – Molecular Glue Direct service takes advantage of our unique capability for multiplexing. POI(s) and E3 ubiquitin ligase(s) are screened in a multiplexed format and compared to the monoplexed screens. Molecular glue hits are observed as 1) hits observed both on POI and E3 ubiquitin ligase in the multiplexed screen, but not on both in the monoplexed screens, and 2) hits only observed in the multiplexed screen but not in the monoplexed screen.
Service in brief
- 1. You provide the purified target protein(s) and E3 ubiquitin ligase(s) (micrograms)
- 2. We conduct a feasibility study and provide a report
- 3. You say ”go”
- 4. We conduct the screening, perform the analysis and provide a report, including hit list with chemical structure information
Timelines
Feasibility study: 4 weeks
Screening: 4-8 weeks
Keys
-
Screen requires small quantities of purified target protein(s) and E3 ubiquitin ligase(s) (micrograms)
-
Screen is conducted multiplexed: POI(s) and E3 ubiquitin ligase(s)
-
Screen is conducted with many conditions in parallel
-
Screen is conducted in a homogeneous assay to avoid matrix binders and target denaturation
-
Identifies molecular glue directly
-
Identifies hits and hit families
-
Instant structure-activity relationship (SAR)
-
Success rate 25-30%
-
To mitigate the relatively low success rate most molecular glue projects have either multiple targets or multiple E3 ubiquitin ligases
-
Applicable across disease areas and target classes (no structural information required)
-
Straightforward and transparent data analysis
Business Model
- Simple fee-based plan with no reach-through and royalty payments
- Hit exclusivity
- The compensation plan comprises a total of 3 payments
- Pre-Project Feasibility Study Fee – scales with work
- Screening Fee – scales with screening slots
- Chemical Series Nomination Fee
- The project flow is rapid and straightforward:
- Client provides the target protein(s) and E3 ubiquitin ligase(s) (100 micrograms)
- Vipergen performs Feasibility Study (go-no-go)
- Vipergen performs two rounds of screenings in monoplexed and multiplexed formats, each under varying conditions and with different libraries.
- After each screening round, Vipergen delivers a hit list and report, including metrics from screens and chemical structure information
- Hits are reserved for client for 1 year
- Client may resynthesize and test hits in relevant assays and use data for 1 year
- Client may nominate Chemical Series for 1 year
- Vipergen assigns its rights to Nominated Chemical Series and these become exclusive for client
DELs
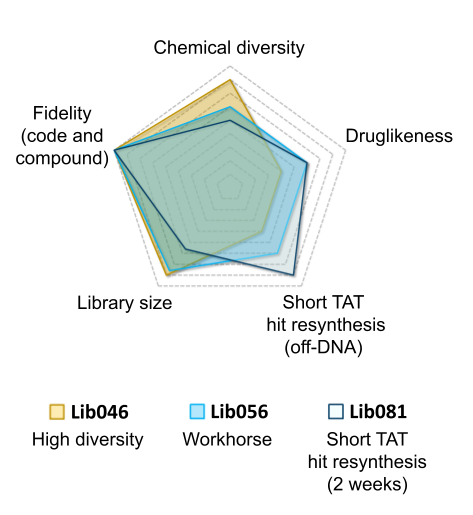
Vipergen’s high fidelity DELs are synthesized using robust chemistry and with a purification step after each chemical transformation, thus providing 100% correspondence between code and compound (no truncates).
Key drivers for library design
- Chemical diversity
- Physicochemical properties
- Fidelity
- Hit resynthesis (off DNA)
| Parameter | Lib046 | Lib056 | Lib081 |
|---|---|---|---|
| Size (million of compounds) | 445 | 522 | 381 |
| Chemistries |
|
|
|
| Compounds w 1 cyclic, tertiary amide | 49% | 47% | 52% |
| Compounds w 2 cyclic, tertiary amides | – | 42% | 33% |
| Total number of building blocks (#) | 1507 | 1327 | 1138 |
| Designer building blocks (#) | 979 | 755 | 617 |
| Scaffolds (#) | 368 | 343 | 295 |
| Toxicophores (%) | 1.1 | 0.4 | 1.1 |
| Molecular weight (avg) | 612 | 529 | 525 |
| cLogP (avg) | 2.8 | 0.75 | 0.9 |
| HBA (avg) | 8.7 | 6.3 | 6.2 |
| HBD (avg) | 2.8 | 2.8 | 2.9 |
| Rotatable bonds (avg) | 10.7 | 8.8 | 8.9 |
| TPSA (avg) | 153 | 139 | 137 |
| Fsp3 (avg) | 0.5 | 0.6 | 0.6 |
Target classes
- Kinases
- Oxidoreductase Proteases
- Hydrolases
- Isomerases
- Transferases
- Lyases
- Phosphatases
- Transcription factors
- Synthases
- E3 biquitin ligases
- Deubiquitinases (DUBs)
- Helicases
- Polymerases
- Nucleotide exchange factors
- Decarboxylases
- Ligases
- Integral membrane proteins
- G protein-coupled receptors (GPCRs)
- Ion channels
- Single-pass membrane proteins
- Nuclear receptors
- Cytosolic receptors
- Extracellular receptors
- Transcription factors
- E3 ligases
- Deubiquitinases (DUBs)
- Antibodies
- Cytokines
- Interleukins
- Growth factors
- Carrier proteins
- Adaptor proteins
- Oncoproteins
- Histone binders
- Methyl transferases
- Acetyltransferase
- Demethylases
- Deacetylases
- Viral proteins (enzymes and coat proteins)
- Bacterial proteins (enzymes and receptors)
- Fungal proteins (enzymes and receptors)
Technical information
Related Services
Small molecule drug discovery for even hard-to-drug targets – identify inhibitors, binders and modulators | |
Molecular Glue Direct | |
PPI Inhibitor Direct | |
Integral membrane proteins | |
Selectivity Direct – multiplexed screening of target and anti-targets | |
Express – optimized for fast turn-around-time | |
Snap – easy, fast, and affordable |
