
Tap into the potential of molecular glue with our unique drug discovery technology – partner with Vipergen
DELs in Cells – Molecular Glue Direct
The DELs in Cells – Molecular Glue Direct service offers a unique method for directly discovering molecular glues for your target protein
Molecular Glue Direct service in brief:
-
Provide target protein sequence: You provide the amino acid sequence(s) of the target protein(s) of interest (POI).
-
Injection, co-expression, and screening: We inject mRNA encoding the POI(s) and E3 ubiquitin ligase(s) into living cells for co-expression. Screening in a multiplexed format allows for direct discovery of molecular glue hits that induce the interaction between the POI and the E3 ligase.
-
Data report: You receive a data report on hits to advance your drug discovery within a timeframe of less than three months.
The DELs in Cells – Molecular Glue Direct service provides an excellent choice for targets, where targeted protein degradation is the preferred pharmaceutical mode of action.
Do you have an inquiry?
From straightforward Fee-For-Service to comprehensive joint ventures, we provide flexible partnership structures to align with your project’s distinct needs.
-
Innovative screening technologies with Cellular Binder Trap Enrichment (cBTE) & Binder Trap Enrichment (BTE)
-
Vipergen’s approach enables the discovery of novel molecules directly in living cells, improving hit rates and drug development efficiency.
-
Vipergen owns exclusive rights to its technology platforms, including YoctoReactor®, BTE, and cBTE, all secured by strong patent positions
Molecular glues in targeted protein degradation
Unlocking the potential of targeted protein degradation through the cellular ubiquitin-proteasome machinery represents a paradigm shift in drug discovery, broadening the scope of druggable targets. This process involves stabilizing a complex between the POI and one of the 600+ E3 ubiquitin ligases encoded by the human genome, promoting POI ubiquitination and subsequent degradation.
Molecular glues hold promise in this field owing to their mechanism of action and their physicochemical properties, but the discovery of such molecules has proven challenging by traditional methods.
Our solution – DELs in Cells – Molecular Glue Direct, is a unique technology to directly screen for molecular glues in living cells, offering our partners access to a fast and efficient screening format.
Screening for molecular glues
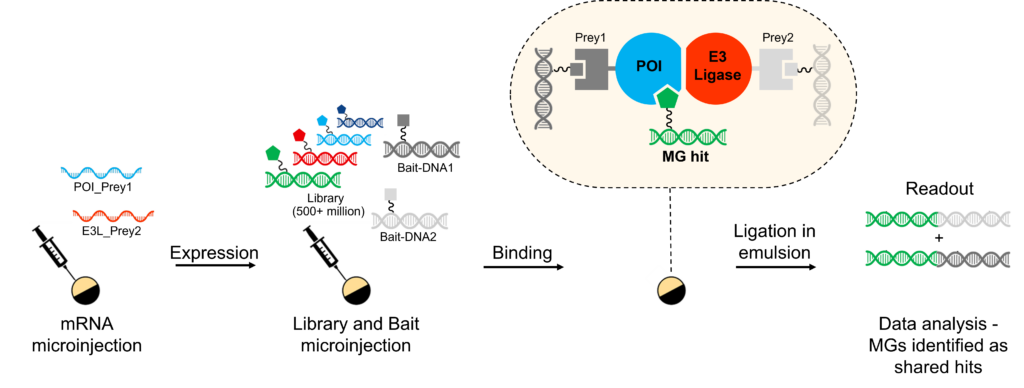
DELs in Cells – Molecular Glue Direct enables discovery of molecular glue molecules by screening the POI(s) in a living cell, using our cellular Binder Trap Enrichment (cBTE) technology in our unique multiplexed format.
The multiplexing capability allows for the simultaneous screening of multiple proteins, including a POI and an E3 ligase, in the same living cell. This is achieved by tagging each target protein with barcoding DNA, and using the barcode to enable the discovery of compounds binding to the POI, the E3 ligase, and the complex between them, respectively.
In the subsequent analysis, molecular glues are identified as the molecules that are found as shared hits between the POI and the E3 ligase.
The multiplexing cBTE technology can also be used for identifying molecular glues in more general cases, where the interaction partners do not include an E3 ligase, and can even be used to identify inhibitors of protein-protein interaction directly.
Off-the-shelf human E3 ligases
-
Cereblon
-
DCAF15
-
DCAF11
-
DDB1
-
KEAP1
-
KLHDC3
-
VHL
-
PELI1
-
MDM2
-
TRIM25
-
PRAJA1
-
PRKN
-
UBE3A
-
CHIP
Streamlined discovery of molecular glues
DELs in Cells – Molecular Glue Direct facilitates the discovery of molecular glues without the requirement of purified target proteins, as the targets are expressed as an intrinsic part of the project.
Projects are run in two phases:
Feasibility study
- Design of constructs
- Expression of the POI(s) in Xenopus oocytes
- Co-expression with E3 ligases (off-the-shelf or your preferred)
- Evaluation by Western blotting (POI size and level of expression)
- Enables screening for molecular glues by achieving balanced expression of POI and E3 ligase.
Human E3 ligases are active in Xenopus oocytes, so upstream ubiquitination inhibitors are included during expression to prevent target ubiquitination and degradation.
Our scientists have a strong history of successfully expressing target proteins in Xenopus oocytes, with over 95% of projects succeeding at this stage.
Multiplexed DEL screening
- Balanced expression of POI and E3 ligase(s) in Xenopus oocytes
- Upstream ubiquitination inhibitors included to prevent target ubiquitination and degradation
- DEL and Bait-DNAs are injected into the cell
- Binding is encoded by ligation in emulsion
- Hits are identified by DNA sequencing and decoding
For projects where there’s an inherent affinity between the POI and the E3 ligase, we employ a more advanced method called off-rate based screening to identify molecular glues. This method exploits the increased lifetime of the POI-E3 ligase complex upon binding of the molecular glue.
After hits are identified through DNA sequencing and decoding, they are grouped into clusters based on their structural similarity and reported to the partner. Our data analysis effectively distinguishes between compounds binding to the target protein alone and those binding to the target protein complexed with the E3 ligase, facilitating the discovery of molecular glues.
How is direct screening for molecular glues enabled?
Our team of molecular biologists and chemists are experts in screening technologies and have a strong track record of delivering fast and reliable results. We have optimized DEL screening in living cells to enable direct screening for molecular glue.
The drug target protein is expressed as an intrinsic part of the process
With DELs in Cells – Molecular Glue Direct, you only need to provide the amino acid sequence of the target protein(s). The drug target protein expression is performed as an intrinsic part of the process, eliminating the need for purified target protein.
The drug target protein is screened in a multiplex setup
Data analysis
Case study: Discovery of molecular glues for an oncology target
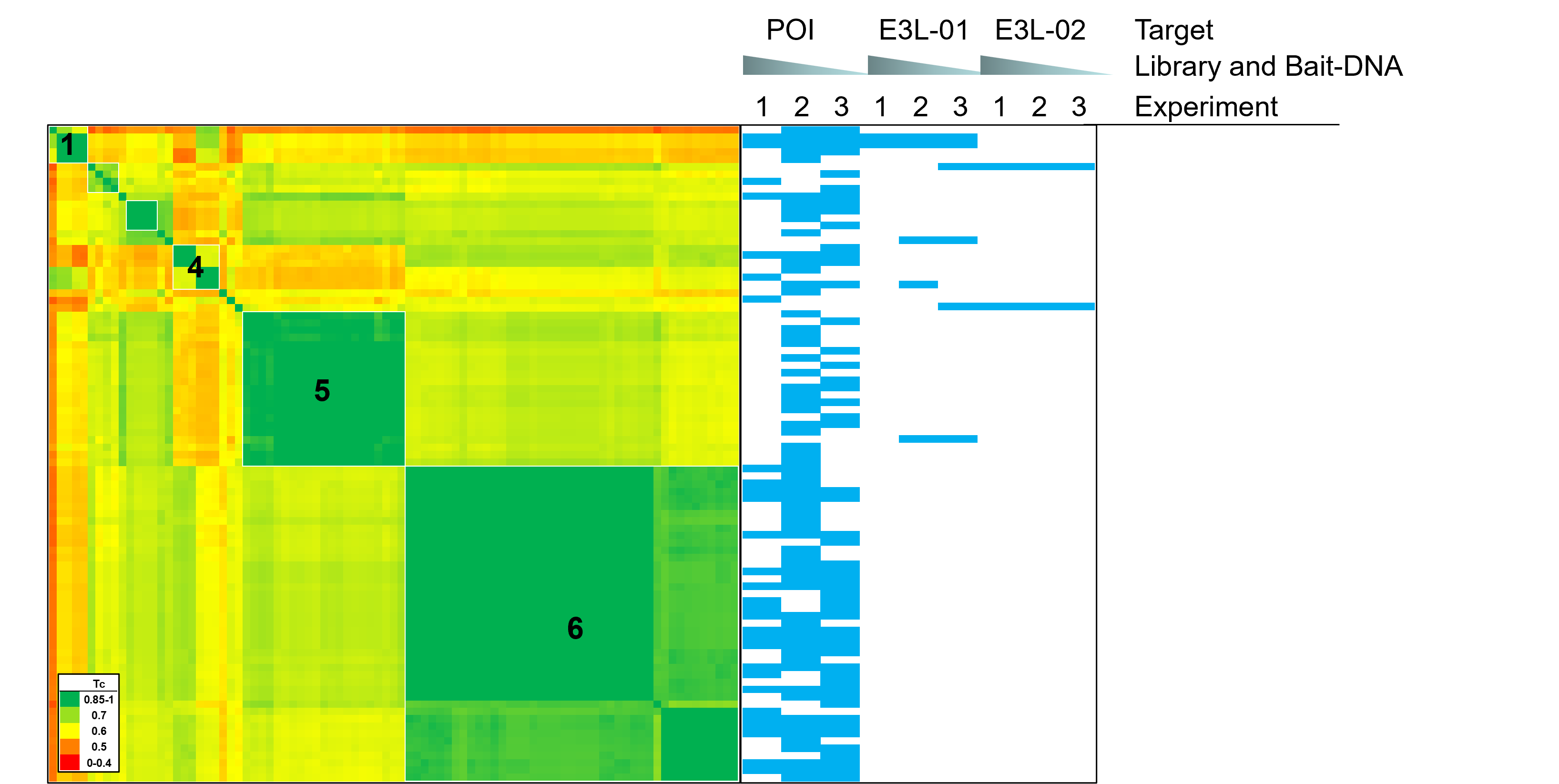
Result of a DELS in Cells – Molecular Glue Direct screen
- An oncology target was expressed in triplex together with two E3 ligases in a DELs in Cells – Molecular Glue Direct screen
- 89 hits were discovered
- Hits were clustered in 6 chemical series by pairwise Tanimoto similarity
- Cluster 1 and 4 contain hits shared between the POI and E3L-01
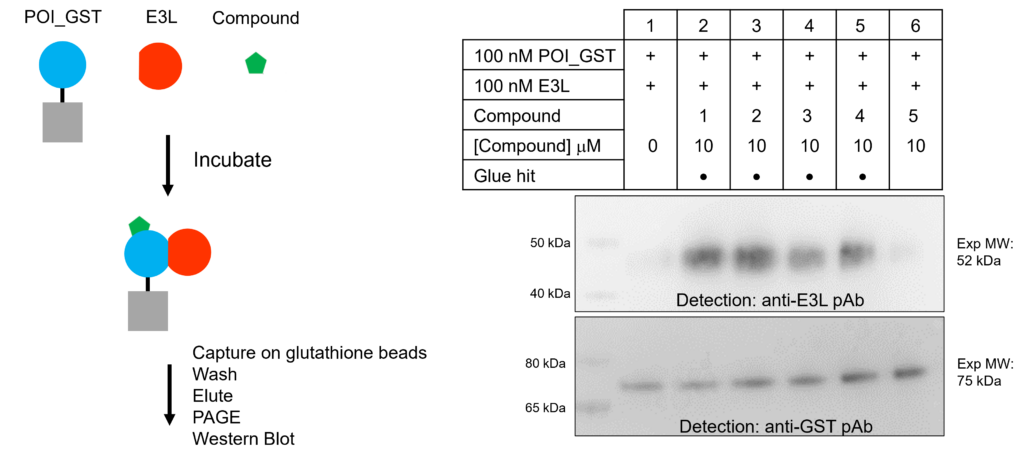
Validation of molecular glue compounds in a Co-IP assay
- Interaction between the POI and E3L-01 was analyzed in a co-immunoprecipitation (Co-IP) assay
- Four out of four compounds discovered induced the interaction between the POI and the E3L-01
- One unrelated compound did not induce the interaction
Conclusion
Molecular glue compounds were discovered in a DEL in Cells – Molecular Glue Direct screen and validated in a secondary assay.
Frequently Asked Questions
PROTACs are heterobifunctional molecules that induce ternary complex formation between the target protein and E3 ligase through two distinct small molecule–protein interactions. Molecular glues, on the other hand, are small molecules that either induce interaction or enhance an existing affinity between a protein pair through cooperative binding.
The mechanism of binding renders PROTACs susceptible to the “hook effect,” where high concentrations of PROTACs lead to decreased efficacy due to binding site saturation.
Overall, PROTACs are generally easier to discover compared to molecular glues, but their development is more challenging due to their larger size and complex physicochemical properties.
No, unlike other screening technologies, only the amino acid sequence of the target protein is needed to execute the screening project, enabling a rapid progression from project initiation to hits that provides our partners with a competitive edge.
The target protein expression is ensured by Western blotting, a rapid technology enabling semi-quantitative and qualitative assessment of the expressed protein’s integrity.
