
YoctoReactor Libraries
Chemical building blocks (BBs) are brought into close proximity which facilitates chemical reaction and the attached DNA ultimately encodes the final product.
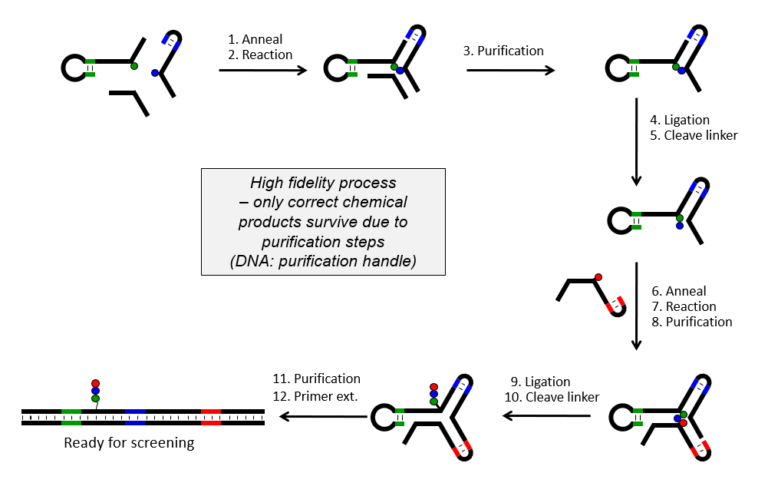
The overall design has favorable implications for the scope and reliability of the chemistry, the stability of the structure, and, ultimately, the capability for addressing the most challenging targets. 100% correspondence between code and synthesized compound is ensured by purification steps using the DNA as purification handle after each chemical step.
A fundamental difference between conventional HTS screening and DEL, is that hits identified by DEL screens need to be resynthesized off DNA. Therefore, we have a particular focus on a low false positive rate.
Library Philosophy
- Fidelity
- Make-a-bond purify/break-a-bond purify – no truncated products
- Robust chemistries
- Chemical diversity
- Diverse sets of building blocks (main contributor)
- Few chemistries
- Physicochemical properties, especially keep cLogP and MW in check
- 3 diversity points
- Universal libraries (no target class focused)
Combinatorial assembly of vast DNA-encoded small molecule libraries
- A library consists of hundreds of millions of DNA-encoded small molecules formed by combinatorial self-assembly of DNA-encoded BBs
- The identity of the product is easily determined by DNA sequencing
- The yR affords precise control of chemical reactivity and deconvolution
Chemical diversity – building block generated
The chemical building blocks (BBs) are responsible for the vast majority of atoms and bonds in the library compounds. Consequently, BBs represent our primary focus for generating chemical diversity. We use commercial available BBs when possible, but are using in-house designed BBs in increasing numbers to satisfy the need for generating interesting and novel diversity.
- More constrained structures
- More complex structures
- Less rotatable bonds
- Novel heterocyclic motifs
- More tertiary amides (better solubility, membrane permeability, plasma and metabolic stability)
100% match between code and compound – stepwise yR library synthesis
yR library example – Lib056
- Type – Trimeric format
- Size – 522 million (1 compound – 1 unique DNA code)
- Synthesis – Robust and reliable: 2 different chemistries in 2 steps: acylation and reductive amination
- Novel compounds – High: 99.9% with a Tanimoto similarity score < 0.95 with ChEMBL
- Toxicophores – Low: ⁓0.4%
- Oral bioavailability – High: 99.2% in compliance with Ro5 (MW 529 Da and cLogP 0.75 on average)
- Aliphatic content – High: Fsp3 = 0.6 on average (good for solubility and provides 3D structures)
- Diversity – High: Driven by BBs – carefully designed and chosen for pharmacophore motifs, 3D orientation and even grid-size
- Building Blocks – High:
- 1327 different BBs
- 795 designer BBs
- 572 commercially available BBs
- 1327 different BBs
- Scaffold diversity – High: 341 different
- Topological diversity – balanced:
- Rigidity/flexibility: 0.45 on average (rigid<0.5<flexible)
- Shape index: 0.59 on average (Spherical<0.5<linear)
- 3D optimized
- Building Blocks – High:
Why Bigger Isn’t Always Better: Choosing the Right DNA Encoded Library Size for DEL Screening
DNA-encoded library (DEL) technology dazzles drug-hunters with the promise of “billions and billions” of compounds in a single tube. Rapid one-pot affinity selections, parallel screening under multiple conditions and deep sequencing all contribute to its appeal. Yet the gravitational pull of sheer scale has encouraged many providers to trumpet ever-larger libraries as a competitive badge of honour, and decision-makers routinely benchmark vendors by headline numbers alone. Beneath the surface, however, the connection between numeric size and real-world screening success is far less linear than marketing suggests.
The Intuitive Appeal – and Hidden Pitfalls – of Ultra-large DELs
On paper, e.g. jumping from 400 million to 10 billion library members looks like a 25-fold improvement in chemical space. In practice, multiple intertwined factors erode that advantage:
- Sampling depth limits: A sequencing run delivers a cap on number of reads. When those are spread across billions of barcodes, each compound is sampled so sparsely that genuine binders might disappear in statistical noise, demanding extreme enrichment to stand out as a hit.
- Copy numbers economic: To keep more than 10^6 copies of each library member (Empirically determined minimum for reliable hit recovery [Satz 2017]) you would quickly need grams of material; a 10^12 member DEL would require ~1.6 µmol for a single selection.
- False-negative inflation: Meta analysis of industry data shows that larger numeric libraries suffer higher miss rates because potentially useful mid-affinity ligands are lost below the detection threshold.
The conclusion: After library sizes in the mid-hundreds of millions of well-behaved library members, the potential advantages diminish rapidly with further growth (Wichert 2024).
High-Fidelity Beats High-Numeracy
After all, chemistry matters. Every additional synthetic cycle multiplies the risk of truncates, side-products and DNA-damage. This in the end dilutes the proportion of correctly encoded molecules. High-yielding reactions, careful building-block vetting and robust QC create faithful libraries where each barcode genuinely represents the intended unique, druglike structure. Such fidelity improves downstream hit validation rates. At Vipergen we have over the years curated our yoctoReactor (Hansen 2009, Blakskjaer 2015) technology, which allows for the construction of DELs without the presence of any truncates yielding a 100% match between barcode and displayed small molecule.
Chemical diversity over redundancy
A smaller library made from judiciously balanced building blocks across various heterocycles, three-dimensional shape, polarity and Fsp3 will cover more biologically relevant chemical space than a large library of flat aromatic molecules.
Studies comparing focused, physiochemically balanced DELs with vast random DEL mixes shows no loss – and often a gain – in ligand discovery efficiency (Chen 2020). Likewise, Eidam and Satz demonstrated that reaction‑validated libraries of 10^8–10^9 members recovered more high‑quality leads as 10^11 counterparts while producing fewer false negatives (Eidam & Satz 2016).
Key elements of diversity include:
- Scaffold and exit vector variety
- Stereo- and sp3-rich motifs
- Drug-like size (3-500 Da) and lipophilicity (cLogP < 4)
- Privileged pharmacophore coverage
- Reduction of PAINS, matrix binders and frequent hitters
Cost, speed and sustainability
Sequencing costs keep falling, but protein, reagents, and lab time still dominate project budgets. Handling kilogram-scale bead slurries, PCR of picogram DNA and multi-round selections on fragile target proteins all add complexity. By utilizing a right-sized DEL can:
- Running more conditions (mutants, counter-targets, competition experiments) per project at equal cost
- Iterate library design faster
- Reduce environmental footprint through lower chemical consumption
References
- Blakskjaer, P. et. al., Fidelity by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery, Curr. Opin. Chem. Biol., 2015, 26, 62-71. doi.org/10.1016/j.cbpa.2015.02.003
- Chen, Q. et. al., Exploring the Lower Limit of Individual DNA-Encoded Library Molecules in Selection, SLAS Discovery, 2020, 25, 5, 523-539. doi.org/10.1177/2472555219893949
- Eidam, O. and Satz, A., Analysis of the productivity of DNA encoded libraries, Med. Chem. Commun., 2016, 7, 1323-1331. doi.org/10.1039/c6md00221h
- Hansen, M. H. et. al., A Yoctoliter-Scale DNA Reactor for Small-Molecule Evolution, J. Am. Chem. Soc., 2009, 131, 1322-1327. doi.org/10.1021/ja808558a
- Satz, A. et. al., Analysis of Current DNA Encoded Library Screening Data Indicates Higher False Negative Rates for Numerically Larger Libraries, ACS Comb. Sci., 2017, 19, 4, 234-238. doi.org/10.1021/acscombsci.7b00023
- Wichert, M. et. al., Challenges and Prospects of DNA-Encoded Library Data Interpretation, Chem. Rev., 2024, 124, 22, 12551-12572. doi.org/10.1021/acs.chemrev.4c00284
