Phage Display Technology: Principles, Libraries, and Applications in Drug Discovery
Introduction
Phage display technology is a molecular screening method in which bacteriophages are engineered to display peptides or proteins on their surfaces while carrying the encoding DNA internally. This direct genotype–phenotype linkage enables the construction of phage display libraries with up to 10¹⁰ variants, allowing high-throughput screening for molecules with desirable binding properties. The concept was pioneered in 1985 by George P. Smith, who demonstrated that foreign peptides could be fused to phage coat proteins and displayed in a heritable fashion.
Since its introduction, phage display has become a cornerstone of molecular discovery, particularly in the fields of antibody engineering, peptide ligand discovery, epitope mapping, and vaccine development. The technology’s ability to generate human therapeutic antibodies has had profound impact, with adalimumab (Humira) standing as a landmark example of a phage display–derived monoclonal antibody approved for clinical use [1,2].
Despite the emergence of other display and library technologies such as mRNA display [3,4] and DNA-encoded libraries (DELs) [5-8], phage display remains highly relevant due to its robustness, scalability, and direct compatibility with therapeutic antibody pipelines. In this review, we examine the principles, methods, and applications of phage display technology, with extended comparisons to mRNA display and DEL platforms.
How Phage Display Works
Genotype–phenotype linkage
The foundation of phage display technology is the physical coupling of genotype (encoding DNA) with phenotype (displayed peptide or protein). A foreign DNA sequence is inserted into the gene encoding a phage coat protein, resulting in the fusion of the encoded peptide/protein to the coat protein displayed on the viral surface. The corresponding DNA resides inside the phage particle, ensuring that each displayed molecule is linked to its genetic blueprint.
Common phage systems
Several bacteriophage systems are employed in phage display:
- M13 filamentous phage: The most widely used system. The minor coat protein pIII (3–5 copies) is suited for displaying larger proteins such as antibody fragments, while the major coat protein pVIII (~2700–3000 copies) supports high-valency display of short peptides.
- T7 phage: Offers greater robustness and can display larger proteins without requiring secretion through the bacterial membrane.
- T4 phage: Capable of displaying very large proteins and multivalent constructs.
- λ phage: Less common but useful for certain protein formats.
Helper phages and phagemid vectors are often employed in M13 systems. Phagemids carry the display construct, while helper phages provide the structural proteins needed for phage assembly.
Biopanning cycle
The process of phage display screening, also known as biopanning, involves iterative rounds of affinity selection:
- Library incubation – A large phage display library is exposed to an immobilised target (purified protein, peptide, or even whole cells).
- Washing – Non-binding phages are washed away under increasingly stringent conditions.
- Elution – Bound phages are eluted, often by pH shift, enzymatic cleavage, or competitive ligands.
- Amplification – Eluted phages are amplified in E. coli, regenerating the pool for the next round.
- Enrichment – After 3–5 rounds, high-affinity binders are enriched and sequenced.
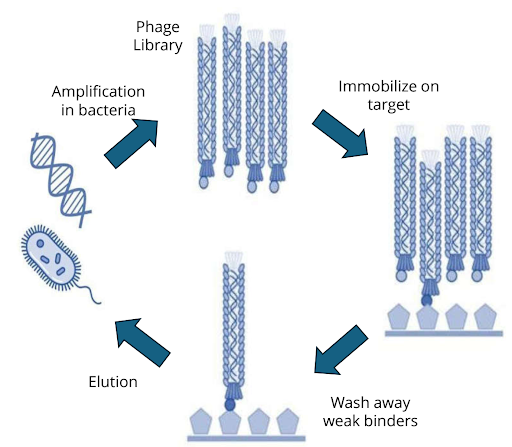
Figure 1: Biopanning cycle
What Is a Phage Display Library?
A phage display library is a diverse collection of bacteriophages, each presenting a unique peptide, antibody fragment, or protein variant on its surface. The diversity of a library can reach 10⁹–10¹⁰ unique members, significantly enhancing the probability of identifying rare, high-affinity binders.
Types of phage display libraries
- Peptide libraries:
- Synthetic libraries use randomised oligonucleotides to generate peptides with controlled diversity.
- Natural libraries use DNA fragments from biological sources.
- Semi-synthetic libraries combine both strategies.
- Antibody libraries:
- Naïve libraries are derived from B-cell repertoires of healthy donors, constructed via splice-by-overlap extension PCR, providing broad diversity.
- Immune libraries originate from immunised donors and typically yield higher-affinity clones against the specific antigen.
- Synthetic antibody libraries incorporate designed CDR (complementarity-determining region) sequences.
- Fragment formats:
- scFv (single-chain variable fragments)
- Fab (fragment antigen-binding)
- VH and nanobody formats
Vector systems
- Type-3 vectors fuse peptides to all copies of pIII.
- Type-33 vectors allow wild-type and fusion proteins on the same phage.
- Phagemids combined with helper phages enable modular display with controlled copy number.
Applications of Phage Display
Phage display technology has broad applications across biomedical research and drug discovery:
Antibody engineering
Peptide ligand discovery
Phage display peptide libraries are valuable tools for identifying binding motifs against receptors, enzymes, and protein–protein interaction interfaces. Short peptide ligands discovered via phage display have been used as leads for therapeutics, diagnostics, and targeting agents.
Epitope mapping and vaccine design
Epitope mapping with phage display identifies the precise binding sites of antibodies on antigens, aiding rational vaccine design and immunodiagnostic assay development.
Emerging applications
- Nanobody generation: Selection of VH domains and nanobody scaffolds against membrane proteins.
- Cell-surface selection: Direct screening against intact cells for immuno-oncology targets.
- Diagnostic peptides: For example, HER3P1 peptide has been developed for imaging HER3 expression in tumors.
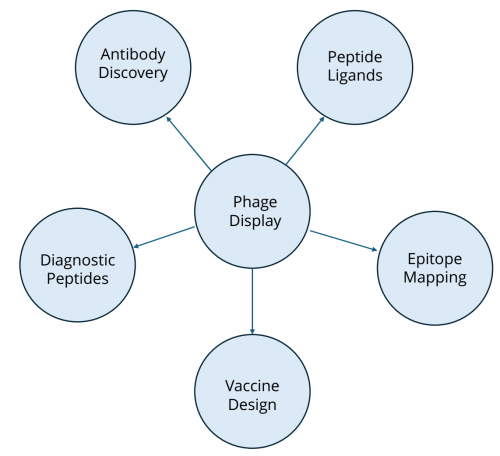
Figure 2: Application of phage display
Advantages and Limitations of Phage Display
Advantages
- Extremely large library sizes (up to ~10¹⁰ variants)
- Direct genotype–phenotype linkage
- Straightforward amplification in bacteria
- Cost-effective and scalable
- Rapid identification of high-affinity ligands
Limitations
- Restricted to peptides and proteins (no small-molecule diversity)
- Post-translationally modified proteins are difficult to display
- Large proteins may not fold correctly in the phage context
- cDNA libraries may contain stop codons or non-functional clones
Comparison with Other Display Platforms
mRNA Display
mRNA display is a cell-free in vitro translation system where peptides are covalently linked to their encoding mRNA via puromycin [3]. Key features include:
- Library size: Up to 10¹³ members, surpassing phage libraries.
- Chemical diversity: Incorporation of some non-natural amino acids and macrocyclic scaffolds [4].
- Applications: Particularly suited for targeting protein–protein interaction (PPI) interfaces.
DNA-Encoded Library (DEL) Technology
DEL combines combinatorial chemistry with DNA barcoding [5-7]. Features include:
- Library size: 10⁶–10¹² small molecules.
- Chemical diversity: Very high; compatible with drug-like small molecules.
- Screening: Pooled libraries incubated with targets, followed by PCR enrichment and NGS readout [6].
- Applications: Small-molecule discovery, especially for enzymatic targets, PPIs, and molecular glues.
- Integration with AI/ML: Machine learning enhances predictive power and hit triaging [8].
| Parameter | Phage Display | mRNA Display | DEL Technology |
|---|---|---|---|
| Molecular class | Peptides, proteins | Peptides, macrocycles | Small molecules and peptides |
| Library size | 109–1011 | 1012–1013 | 106–1012 |
| Chemical diversity | Limited | Moderate to high | Very high |
| In vitro compatibility | Partial | Full | Full |
| Target types | Proteins, cells | Proteins, PPIs | Proteins, enzymatic targets, molecular glues, PPIs |
Table 1: Comparative table of phage display, mRNA display, and DEL
Trends in Commercial Use and Collaboration
Pharmaceutical companies increasingly rely on collaborations with contract research organizations (CROs) and platform providers to access proprietary phage display libraries, custom library construction, and data analysis expertise. Outsourcing offers:
- Access to validated and proprietary antibody/peptide libraries
- Customisation for specific targets (including membrane proteins and GPCRs)
- Data interpretation, clustering, and selectivity profiling
Integration with structure-based drug design, cryo-EM, and computational modelling further accelerates lead optimization. AI-augmented approaches, particularly in DEL screening, are increasingly being applied to phage display datasets as well.
Vipergen’s DNA-Encoded Library Screening Platform
Although this review is centred on phage display, complementary technologies such as DNA-encoded library screening services play a vital role in small-molecule discovery. At Vipergen, we provide custom DEL screening solutions with:
- Modular library design with privileged scaffolds and novel chemotypes
- Screening against challenging targets, including membrane proteins and in intact cells
- Integrated SAR analysis and cheminformatics clustering
- Multiplexed selectivity profiling against target and anti-targets
For researchers pursuing oncology, GPCR ligands, or chemical probes for validation, our DEL screening services provide a high-throughput, scalable platform that complements phage display–based biologics discovery.
Conclusion
Phage display technology remains one of the most influential methods in molecular discovery. By harnessing genotype–phenotype linkage, phage display libraries enable rapid screening of billions of peptides, proteins, and antibody fragments. The method’s impact on antibody therapeutics, peptide discovery, and epitope mapping underscores its continuing relevance, even as complementary technologies such as mRNA display and DEL broaden the molecular space accessible to researchers.
As library construction methods, biopanning strategies, and computational tools evolve, phage display screening will continue to serve as a cornerstone of biologics discovery while integrating seamlessly with next-generation discovery platforms. Its combination of scalability, cost effectiveness, and clinical track record ensures that phage display remains indispensable in the next era of precision drug discovery.
References
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348(6301):552–554. https://doi.org/10.1038/348552a0
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. https://doi.org/10.1146/annurev.iy.12.040194.002245
- Roberts RW, Szostak JW. RNA–peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA. 1997;94(23):12297–12302. https://doi.org/10.1073/pnas.94.23.12297
- Huang Y, Wiedmann MM, Suga H. RNA Display Methods for the Discovery of Bioactive Macrocycles. Chem Rev. 2019;119(17):10360–10391. https://doi.org/10.1021/acs.chemrev.8b00430
- Peterson AA, Liu DR. Small-molecule discovery through DNA-encoded libraries. Nat Rev Drug Discov. 2023;22(9):699–722. https://doi.org/10.1038/s41573-023-00713-6
- Favalli N, Bassi G, Scheuermann J, Neri D. DNA-encoded chemical libraries – achievements and remaining challenges. FEBS Lett. 2018;592(17):2168-2180. https://doi.org/10.1002/1873-3468.13068
- Mason JW, Wang Y, et al. DNA-encoded library-enabled discovery of proximity-inducing small molecules. Nat Chem Biol. 2024;20:170–179. https://doi.org/10.1038/s41589-023-01458-4
- McCloskey K, Sigel EA, et al. Machine learning on DNA-encoded libraries: A new paradigm for hit-finding. J Med Chem. 2020;63(16):8857-8866. https://doi.org/10.1021/acs.jmedchem.0c00452
Related Services
| Service | |
|---|---|
Small molecule drug discovery for even hard-to-drug targets – identify inhibitors, binders and modulators | |
Molecular Glue Direct | |
PPI Inhibitor Direct | |
Integral membrane proteins | |
Specificity Direct – multiplexed screening of target and anti-targets | |
Express – optimized for fast turn – around-time | |
Snap – easy, fast, and affordable |


